The purpose of this package is to allow a user to have easy access to a consortium of publicly available chemometrics datasets.
Chemometrics data is widely dispersed, in a variety of file formats, and arbitrary conventions. For people wishing simply to load in some data, learn the field, and try new techniques, run a study, or even do a metanalysis this can complicate matters. To ameliorate this, this package attempts to bundle everything into an intuitive and Julia ready format that is ready for investigation or personal exploration.
Data that is obtained or made accessible via this API is either "curated" or "scraped" from public domain resources. As a user it is your duty to investigate the use, utility, and integrity of this data for your purposes or end goals. This package does it's best to inform you of these nuances, but may have flaws in the information available, and even then - sometimes that information changes over-time. See the Liability section for more information.
The code base provides some pretty basic commands to let users kind of poke around for data. Maybe you don't know what dataset you want, but you're interested in trying your hand at some chemometrics tasks using mid-infrared spectroscopy. You could do the following:

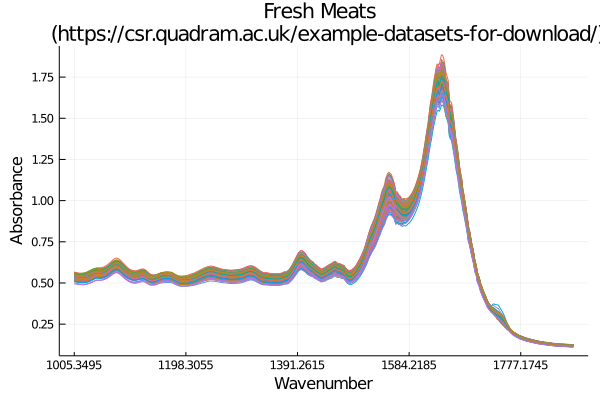
Of course you can easily crack open the datasets and have them ready to use in a few short lines of code. Below is an example workflow of loading in some data, using some metadata, and generating a plot:
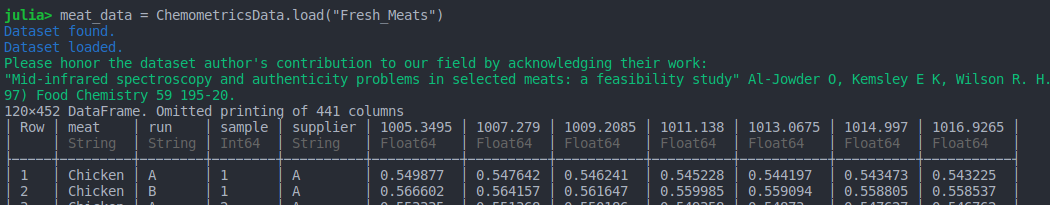
quadrum_data = load("Fresh_Meats")
meta_data = meta("Fresh_Meats")
spectra = numeric_columns( gimme_the_data )
bins = parse.( Float64, names(spectra) )
bin_display_range = 1:100:length(bins)
using Plots
plot( spectra |> Matrix |> transpose,
legend = false,
title = "Fresh Meats \n (" * meta_data["URL"] * ")",
xlabel = "Wavenumber", ylabel = "Absorbance",
xticks = ( bin_display_range, bins[bin_display_range] )
)When a dataset is loaded using this package a reminder is displayed for the user to honor it's provenance in derivative works and publications,
 Authors/owners of datasets can also request custom messages to be displayed. For example, the penicillin dataset will emit the following message on loading: "Please acknowledge the following paper if utilizing the spectral data which can be freely downloaded at www.industrialpenicillinsimulation.com. Goldrick S., Duran-Villalobos C., K. Jankauskas, Lovett D., Farid S. S, Lennox B., (2019) Modern day control challenges for industrial-scale fermentation processes. Computers and Chemical Engineering.".
Authors/owners of datasets can also request custom messages to be displayed. For example, the penicillin dataset will emit the following message on loading: "Please acknowledge the following paper if utilizing the spectral data which can be freely downloaded at www.industrialpenicillinsimulation.com. Goldrick S., Duran-Villalobos C., K. Jankauskas, Lovett D., Farid S. S, Lennox B., (2019) Modern day control challenges for industrial-scale fermentation processes. Computers and Chemical Engineering.".
There are several ways to contribute to this effort:
- Make a Pull Request and submit a dataset! This should update the manifest.jl file and provide the data in a plaintext manner in a separate folder.
- File an issue on the repository and point to where your data is. Give written permission for it's redistribution and some form of proof that you are the legal copyright owner of the data!
- Track down Casey Kneale, Ph.D. on social media or ICS-L.
- Single datasets should not exceed 10mb. If it does contact us and we will navigate an alternative means of hosting(IE: not github).
- It is highly preferable that the data is in a tabular nonproprietary file format, such as CSV. Proprietary file formats regularly change: companies go out of business, software gets deprecated, etc.
- if the content or the origin of the data is suspect, it will either be not accepted, or removed.
The creators and contributors to this package are not responsible for the outcomes of the use of any data, or code in this repository. This is to be used at ones own risk. Changes to the end users file system may occur with remotely accessed datasets(obtained with the fetchdata() command). This is because this package will unpack them(after checking their MD5 checksum for authenticity). The offline available data should present minimal risks.
Before redistributing any data accessed via this package (either stored in the git repository or via the packages use) ensure that the permissions you have obtained for this data allow for this. The contributors to this package can only offer permissions for data they themselves own. Just because something is "public domain" does not imply it may be used commercially, or redistributed without author's approval.
Should also be noted that it is possible errors exist in these datasets due to them being manipulated(transposed, file formats changed, etc) to get them into a common form. Please report any and all issues - it's greatly appreciated.
- Obtain permissions for as much data as possible
- Collect and clean that data to a common and easy file format.
- Write functionality for downloading remote datasets that are large.
- Write functionality for scraping remote datasets
- Extend to an online database.
- Write an R/python port for this package so users are free to explore Chemometrics data from free tools of their choice.